Parallelize the flux calulator
Contents
Parallelize the flux calulator¶
It is possible to run the flux calculator in parallel where each process takes care of a specific part of the exchage grid. These parts are determined according to the ocean model’s parallelization: for each process of the ocean model, the corresponding ocean grid cells are identified and exchange grid cells that are linked to these cells are grouped together. Since some fields have to be regridded to other grids (e.g from the t to the u grid) there are overlaps between the groups, these overlaps are called halos, see Fig. 8
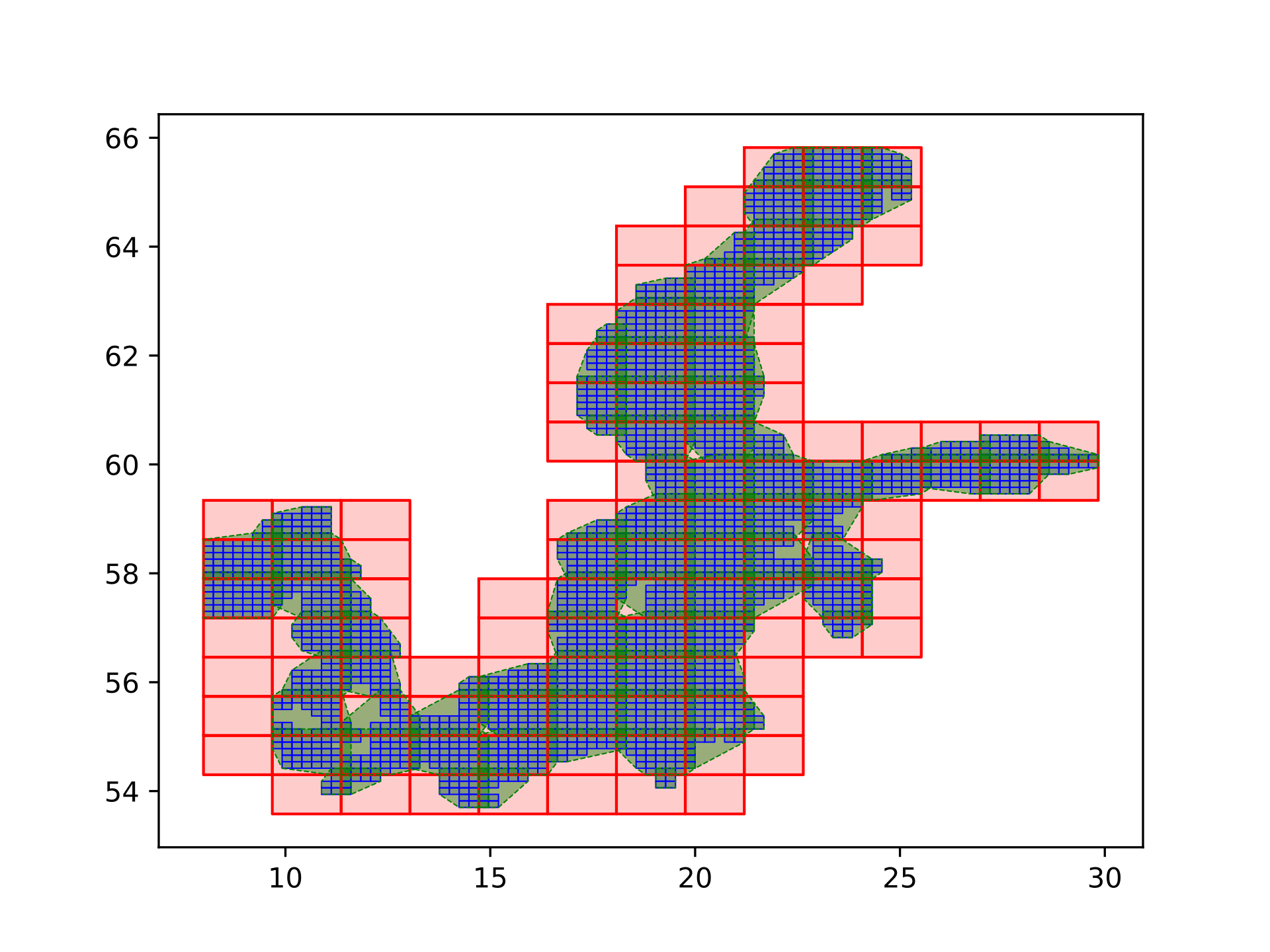
Fig. 8 Domain decomposition for parallelizing the flux calculator. Blue are the ocean model’s grid cells, red is the domain decomposition for the ocean model’s processes and green is the corresponding exchange grid decomposition.¶
Adapt the global settings¶
There are two posibilities to parallelize the flux calculator.
On bottom model cores¶
In order to run the flux calculator processes on the same cores as the ocean model you have to specify
flux_calculator_mode = "on_bottom_model_cores"
in your global_settings.py in the input folder.
Importantly, if you use Intel MPI for paralleization on the HLRN machines you have to put
export PSM2_MULTI_EP=0
into your jobscript template after the MPI module has been loaded. This enables putting more tasks on the node than available cores, see also https://www.hlrn.de/doc/display/PUB/MPI+Jobs+with+more+than+40+(96)+tasks+per+node+failing.
On extra cores¶
In order to run the flux calculator processes on extra cores you have to specify
flux_calculator_mode = "on_extra_cores"
in your global_settings.py in the input folder.
This option yields the shortest computation time but consumes more computational resources since the flux calculator and the ocean model run on different cores.
Create mappings¶
If you want to switch between the single and parallel mode of the flux calculator, you have to execute the create_mappings.py script in scripts/prepare.
You must do that before you can run the model and after you have set the desired mode in the global_settings.py.
You can execute it via the shell on the target machine
cd scripts/prepare
python3 create_mappings.py
Alternatively you can use the prepare-before-run option of the run script, see Prepare mappings before the run starts.
Techinically the parallelize_mappings.py script in scripts/prepare creates the mapping files that are necessary for the parallel mode.
Note that if you change the domain decomposition of on of your bottom models you have to execute the parallelize_mappings.py script again.